A Monohybrid Cross is a cross between two parents that involves only one trait.
For example lets take a cross between a heterozygous mother and father for the height trait.
Tall = T short = t
In the end you would use a punnet square, and cross the genotypes.
You would end up with a 25% chance for a genotype offspring of TT, 50% chance for a genotype offspring of Tt, and 25% chance of a genotype offspring of tt.
You would have 75% chance of having a phenotype/appearance of a tall kid, and 25% for a shortie, like me.
DiHybrid Cross is when you cross two parents and you are mixing two traits.
For example you are mixing a mother who is heterozygous round and heterozygous yellow. (RrYy) and a father who is the same.
R=round seed, r=wrinkled seed, Y= yellow, y= green
First you would have to come up with all possible combinations in the genotypes of each parent.
Eg. RrYy, if you chose two letters at random, could be RY, rY, Ry, or ry.
so you match those 4 options with the 4 you get from the father and you end up with a possibility from 16 offspring.
In this case the resulting genotype ist 9/16 RRYY, 3/16 RRyy, 3/16 rrYY, and 1/16 rryy
for genotype its 9 yellow round, 3 green round, 3 wrnikled yellow, and 1 wrinkled green.
Thats Monohybrid and DiHibrid Crossing.
Sunday, December 15, 2013
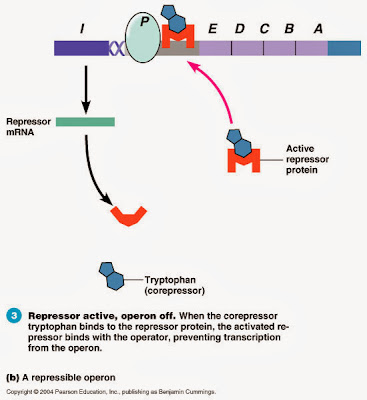
Operon System
DNA Reptile. (Replication)
DNA replication is a process where DNA replicates itself.
There are many enzymes that help in this process.
Helicase- The scissors in the group.
Starting from the Origin of Replication (the starting point) Helicase, (In class we used a yellow triangle to represent Helicase. You can do this if you need some visual aids) breaks down the Hydrogen bonds that connect the two bases.
It does this to unwind the double helix, and prepare a single stranded DNA replication.
There are many enzymes that help in this process.
Helicase- The scissors in the group.
Starting from the Origin of Replication (the starting point) Helicase, (In class we used a yellow triangle to represent Helicase. You can do this if you need some visual aids) breaks down the Hydrogen bonds that connect the two bases.
It does this to unwind the double helix, and prepare a single stranded DNA replication.
Then RNA primase comes on and places rna pairings to the single stranded dna. For example, if you had a strand that read CTAG, the RNA paired with it would be, GAUC, with the Thymine being replaced with Uracil.
It does this to allow DNA Polymerse III the ability to read the strands, because it places rna which has a 3' OH base, which DNA polymerse III can read.
DNA polymerse III then comes in and reads the strand of RNA and creates the complementary nucleotides. Each new base forms phosphodeister bonds.
DNA Polymerse I then replaces the rna with the newly created bases.
Ligase glues all the okazaki fragments together.
DNA Structure
According to Wikipedia, Deoxyribonucleic acid (DNA) is a molecule that encodes the genetic instructions used in the development and functioning of all known living organisms and many viruses.
Basically, DNA is our human code. It makes us, us. Every cell in our body has the same DNA in it.
DNA is usually in a shape of a double helix, and has two strands running in opposite directions. (5' - 3', and 3' - 5')
There are 4 bases, Adenine, Thymine, Guanine, and Cytosine. A goes with T and G goes with C to form base pairs. Each base is connected to the sugar phosphate backbone.
Basically, DNA is our human code. It makes us, us. Every cell in our body has the same DNA in it.
DNA is usually in a shape of a double helix, and has two strands running in opposite directions. (5' - 3', and 3' - 5')
There are 4 bases, Adenine, Thymine, Guanine, and Cytosine. A goes with T and G goes with C to form base pairs. Each base is connected to the sugar phosphate backbone.
Thursday, October 24, 2013
Day 18 DNA!!
1. Explain the significance of Mendel.
Gregor Mendel's experiment of breeding pea plants showed that traits from parents are present in the offspring. His tests supported Darwin's theory of "blending inheritance." It was later proved that the changes in the pea plants were due to the changes in the DNA through generations.
2. Draw the structure of DNA and who discovered this structure.
James D. Watson and Francis Crick discovered the structure of a double helix.
3. Explain each of the five examples of variations that occur to DNA and give an example of each.
a. Point Mutation - a single base pair change. the mutation inactivates a gene for a signaling molecule.
ex. Dogs are moderately more muscular when only one copy of the gene is disabled.
b. Insertion- when multiple base-pair sequences are inserted into a gene.
ex. when 800 base-pair sequences are inserted into a pea produces peas that are wrinkled rather then smooth. The intruding DNA disables a a gene necessary for starch synthesis, affecting the sugar and water content.
c. Gene-Copy number- When you copy errors during cell division to duplicate entire genes.
ex. where the enzyme salivary amylase is copied 10x more in humans than chimpanzees, making humans the better starch digestors.
4. What is evo-devo?
Evo-devo is a subspeciality in evolutionary biology that concentrates on studying the effects of changes in important developmental genes and the role they play in evolution.
5. Make a connection between human migration and the mutation of lactose intolerance.
Gregor Mendel's experiment of breeding pea plants showed that traits from parents are present in the offspring. His tests supported Darwin's theory of "blending inheritance." It was later proved that the changes in the pea plants were due to the changes in the DNA through generations.
2. Draw the structure of DNA and who discovered this structure.
James D. Watson and Francis Crick discovered the structure of a double helix.
3. Explain each of the five examples of variations that occur to DNA and give an example of each.
a. Point Mutation - a single base pair change. the mutation inactivates a gene for a signaling molecule.
ex. Dogs are moderately more muscular when only one copy of the gene is disabled.
b. Insertion- when multiple base-pair sequences are inserted into a gene.
ex. when 800 base-pair sequences are inserted into a pea produces peas that are wrinkled rather then smooth. The intruding DNA disables a a gene necessary for starch synthesis, affecting the sugar and water content.
c. Gene-Copy number- When you copy errors during cell division to duplicate entire genes.
ex. where the enzyme salivary amylase is copied 10x more in humans than chimpanzees, making humans the better starch digestors.
4. What is evo-devo?
Evo-devo is a subspeciality in evolutionary biology that concentrates on studying the effects of changes in important developmental genes and the role they play in evolution.
5. Make a connection between human migration and the mutation of lactose intolerance.
Wednesday, October 23, 2013
Day 17- Traces of a Distant Past
Traces of a Distant Past is all about how DNA gives us a clearer picture of the multimillennial trek from Africa all the way to the tip of South America. It goes on to explain the multiple studies over the differences of human genome from person to person. Almost all DNA, at least 99.9 percent of it, is the same from person to person, but by studying the differences in the last .1 percent, scientists are hoping to uncover clues about the journey our ancestors ventured on from continent to continent.
Only recently have scientists began widening their focus range from only a few isolated stretches of DNA to study hundreds of thousands of nucleotides scattered throughout the genome instead of just the comparisons between let's say East Africans and Native Americans. Scientists are able to follow the breadcrumbs that were left behind by our ancestors.
From the video, "Journey of Man", we learned that Y chromosome gives us an opportunity to follow our migratory heritage back to "Adam," just as earlier work in mitochondrial DNA allowed the identification of Eve the mother of all homo sapiens.
In our Y Chromosomes and our Mitochondrial DNA, scieintists are constantly looking for genetic markers - characteristic patterns of nucleotides - and how they differ from population group to population group.
After piecing this information together, Scientists are then able to piece together a map of the travels our ancestors took to spread throughout the world.
(below)
The difficulties with DNA are that unlike Fossils, the rate of mutation can differ from one strand to another, causing inconsistencies. Yet, fossil remains are too rare, and often incomplete.
To fill in the missing blanks that DNA and fossils can't clearly fill, Scientists use more DNA from microbes that hitch rides on humans. (ie. lice, bacteria, and viruses)
Using this information, Scientists have formulated the Out-of-Africa theory. From 50,000 to 60,000 years ago, our ancestors traveled out of Africa, and eventually overtook the Homo Erectus species and converged into once species. Through interbreeding, the species were able to remain one and the same, making us part Neanderthal.
Then there's the multi-regional theory, which states that over the past 1.8 million years ago, that our ancestors evolved from Asia, Europe, and Africa, and eventually emerged as Homo Sapiens.
The Occasional interbreeding made sure we didn't evolve into multiple species.
There is evidence that supports interbreeding from both theories, many fossilized skeletons of H. Sapiens have features reminiscent of earlier hominids. Scientists estimate that the two genomes are 99.5 percent alike. Yet studies have shown that the Y chromosones in Neanderthals differed from ours.
Scientist would like to use this information and these theories to claim that there is no race. We are all brothers to each other, there is no genetic differences, only geographic gradients.
Friday, September 27, 2013
Day??? - I've been so immersed into my biology studies that I don't know what day this is....
First off, What I did on Day ????. (somehow I'll probably remember what day it is and I'll go back and add the missing ones...)
Today we investigated Common Descent with Molecular Biology.
We were trying to compare the relatedness between organisms by examining the amino acid sequence in the protein, Cytochrome c. By comparing the amino acid sequences between many organisms we were able to determine how closely or distantly organism A was from Organism B.
We discovered that the Horse and Cow had only 3 differences, which makes sense since they look similar, share similar bone structure, walk on 4 legs...etc.
We also discovered that a whale and a moth had 22 differences. Which shocked me, for I thought there would be more of a differences. One was a mammal, while the other was a tiny bug. This just shows how close everyone is... creepy.
Quiz: https://docs.google.com/file/d/0B_nnOJLj_iJAaFVCUWxHelRvM1E/edit?usp=sharing
1. The evidence supports that the Mesonychid was an ancestor of the Basilosaurus, because of anatomical similarities. Both appear to share the "1 bone 2 bone blob" structure."
2. marsupials appear to have began on the continent North America. (e).
3. Bats, Birds, and Dragonflys all share analogous structures, wings. Although they all have wings, with basic similarities, light weight and large length, the underlying structure is completely different. This shows that they have evolved from different ancestral structures. It shows convergent evolution, where the independent evolution of similar features appears in different lineages.
4. The Common Descent lab focused on Amino acid sequences. By comparing the differences in amino acid sequences in cytochrome-c from 20 different species side by side, we were able to observe which species shared similarities and differences, thus allowing us to infer that some species shared near common ancestors. With the Rhesus Monkey and the Human's amino acid sequences there was only 1 difference on amino acid number 66. This shows that the Rhesus Monkey and the Human are very closely related, and have shared a close common ancestor. It also shows that our structures and organs may be similar.
5. Homology is the study of similar structures in different organisms that can show a common ancestor. Some examples would be the 1 bone, 2 bone, blob structure that is found in many organisms. It is found in the Bat, the whale, the Human, the seal, the lion and even the orangutan. This shows that all these species shared a common ancestor at one point. (we are all mammals!)
Today we investigated Common Descent with Molecular Biology.
We were trying to compare the relatedness between organisms by examining the amino acid sequence in the protein, Cytochrome c. By comparing the amino acid sequences between many organisms we were able to determine how closely or distantly organism A was from Organism B.
We discovered that the Horse and Cow had only 3 differences, which makes sense since they look similar, share similar bone structure, walk on 4 legs...etc.
We also discovered that a whale and a moth had 22 differences. Which shocked me, for I thought there would be more of a differences. One was a mammal, while the other was a tiny bug. This just shows how close everyone is... creepy.
Quiz: https://docs.google.com/file/d/0B_nnOJLj_iJAaFVCUWxHelRvM1E/edit?usp=sharing
1. The evidence supports that the Mesonychid was an ancestor of the Basilosaurus, because of anatomical similarities. Both appear to share the "1 bone 2 bone blob" structure."
2. marsupials appear to have began on the continent North America. (e).
3. Bats, Birds, and Dragonflys all share analogous structures, wings. Although they all have wings, with basic similarities, light weight and large length, the underlying structure is completely different. This shows that they have evolved from different ancestral structures. It shows convergent evolution, where the independent evolution of similar features appears in different lineages.
4. The Common Descent lab focused on Amino acid sequences. By comparing the differences in amino acid sequences in cytochrome-c from 20 different species side by side, we were able to observe which species shared similarities and differences, thus allowing us to infer that some species shared near common ancestors. With the Rhesus Monkey and the Human's amino acid sequences there was only 1 difference on amino acid number 66. This shows that the Rhesus Monkey and the Human are very closely related, and have shared a close common ancestor. It also shows that our structures and organs may be similar.
5. Homology is the study of similar structures in different organisms that can show a common ancestor. Some examples would be the 1 bone, 2 bone, blob structure that is found in many organisms. It is found in the Bat, the whale, the Human, the seal, the lion and even the orangutan. This shows that all these species shared a common ancestor at one point. (we are all mammals!)
Wednesday, August 28, 2013
BBlockBonds
On Monday August 26th, Our B block bio class did some handouts!
There were two handouts, one on ionic and covalent bonds, and the other on making molecular models.
I picked the better one: making molecular models.
So, for this handout, we were given a Molecular model kit, some colored pencils, and a periodic table.
We were asked to record the # of atoms, make a model, color the model, and draw a dot structure for each formula. We also needed to know if the compound was polar or non-polar.
Here is some of our work:
Lastly, we have our C6H12O6, otherwise known as Glucose.
Glucose is 6 oxygen atoms each attached to a carbon molecule, with a hydrogen attached to each carbon and to each oxygen. Glucose is a polar molecule, and has a total of 24 atoms.
Through this handout, we were able to visualize atomic bonding, represent atomic bonding using dot diagrams, and predict and classify bonds as either polar or non polar.
There were two handouts, one on ionic and covalent bonds, and the other on making molecular models.
I picked the better one: making molecular models.
So, for this handout, we were given a Molecular model kit, some colored pencils, and a periodic table.
We were asked to record the # of atoms, make a model, color the model, and draw a dot structure for each formula. We also needed to know if the compound was polar or non-polar.
Here is some of our work:
 |
| Our Hydrogen Model (H2) |
Above, is a picture of our H2 Model. In a hydrogen molecule, there are 2 atoms, both hydrogen, who form a covalent bond by sharing their only atoms together. Because both atoms are the same, and that there is no difference in electronegativity, they form a non polar relationship.
 |
| Our Water Molecule (H2O) |
Secondly, this is a picture of our H2O molecule, otherwise known as water. Because oxygen only has 6 valence electrons, it needs two more. So along with the 2 Hydrogen molecules, each sharing one of their electrons, H2O becomes an octet. Water is one oxygen forming 2 covalent bonds with 2 hydrogen atoms.
Unlike the hydrogen bond above, Water is polar due to the two hydrogen atoms.
Glucose is 6 oxygen atoms each attached to a carbon molecule, with a hydrogen attached to each carbon and to each oxygen. Glucose is a polar molecule, and has a total of 24 atoms.
Through this handout, we were able to visualize atomic bonding, represent atomic bonding using dot diagrams, and predict and classify bonds as either polar or non polar.
Saturday, August 24, 2013
Dancing Milk
Hi!
Welcome to my blog!
Today my lab partner and I formed an experiment to test the out hypothesis on the Dancing Milk lab video.
In this video, a drop of dish soap is dropped onto a thin plate of milk, with a couple of food coloring drops on top.
Once the dish soap was dropped, the food coloring began to spread out throughout the milk in a fast reaction.
As a group, Elisa and I formed a hypothesis as to why the food coloring reacted the way it did. We formed a hypothesis that the chemicals in the dish soap broke down the lipids and fat bonds in the milk causing them to move around, which was visible to us due to the food coloring.
We recreated the lab, using the same materials; however, we switched the milk with different types of milk to see if the reaction would happen the same.
In the end,
we noticed that the dish soap for all liquids we tested, 2% milk, whole milk, and skim milk did react the same. Proving our hypothesis that the dish soap broke down all the fat in all of these and caused the food coloring to react the way it did.
In this video, a drop of dish soap is dropped onto a thin plate of milk, with a couple of food coloring drops on top.
Once the dish soap was dropped, the food coloring began to spread out throughout the milk in a fast reaction.
As a group, Elisa and I formed a hypothesis as to why the food coloring reacted the way it did. We formed a hypothesis that the chemicals in the dish soap broke down the lipids and fat bonds in the milk causing them to move around, which was visible to us due to the food coloring.
We recreated the lab, using the same materials; however, we switched the milk with different types of milk to see if the reaction would happen the same.
In the end,
we noticed that the dish soap for all liquids we tested, 2% milk, whole milk, and skim milk did react the same. Proving our hypothesis that the dish soap broke down all the fat in all of these and caused the food coloring to react the way it did.
Subscribe to:
Comments (Atom)
.jpg)